GO Enrichment
This post explains how to use LIS to calculate gene ontology (GO) enrichment.
GO enrichment analysis uses statistical tests to determine if a set of provided genes are statistically different than a comparison set (typically, the set of all genes in the organism), for each of the three main gene ontology aspects.
Gene Ontology (GO) is a classification system that describes three aspects of gene function:
- Molecular Function, describing activities such as catalytic activity or finer-scale activities such as lipoprotein lipase activity;
- Cellular Component, describing cellular components such as the cytoskeleton, or finer-scale locations such as centrosome;
- Biological Process, describing biological programs such as signal transduction, or finer-scale processes such as protein serine/threonine kinase activity.
To calculate GO enrichment at LIS, use the gene-list report function in any InterMine instance. For example, at GlycineMine, enrichment can be calculated for genes from ANY Glycine accession and annotation in GlycineMine.
Here are the steps.
1. Enter a gene list under Analyze a List.
Taking soybean (Glycine spp.) as our example, open GlycineMine, and paste a list of genes in the central box (“Analyze a List”). The list can consist of un-prefixed gene IDs such as Glyma.01G022700, but if that gene exists in multiple assemblies or annotation sets, you will see an intermediate page in which you will be asked to select which genes you want to analyze. Therefore, it is generally best to prefix your identifiers with the following four, dot-separated fields: Genusspecies.Accession.Assemblyversion.Annotationversion.GeneID glyma.Wm82.gnm4.ann1.Glyma.01G022700
Also note that the identifiers should be gene IDs rather than mRNAs; these are typically distinguished by a numeric suffix. That is: use gene Glyma.01G022700 rather than mRNA Glyma.01G022700.1
Try one of the gene lists below:
2. Name and save the list.
You can use the provided name if you wish (based on date and time), or you can give it a more meaningful name. Then click the green “Save a list of 10 Genes” button. (Note that if you register with GlycineMine you will be able to save your gene list so that you can use the same list over and over again on the same tool or use that list on various GlycineMine tools.)
3. Examine the Gene Ontology Enrichment results.
The report page will give descriptive information about each gene; and near the bottom of the page are four reports: “Gene Ontology Enrichment”, “Gene Family Enrichment”, “Pathway Enrichment”, and “Chromosome Distribution”.
In the “Gene Ontology Enrichment” box, be sure to check each ontology aspect that you wich to evaluate:
- biological_process
- cellular_component
- molecular_function
It is common for a set of genes to show enrichment for one aspect and not others.
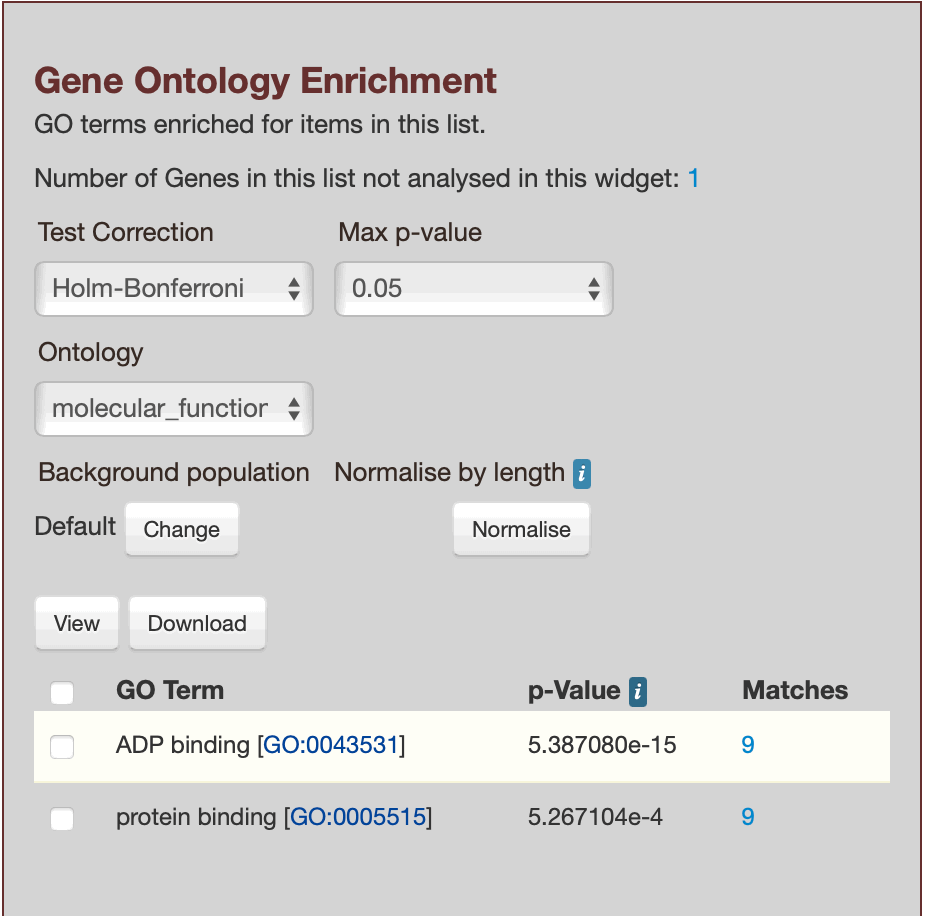
glyma.Wm82.gnm4.ann1.Glyma.01G022700, glyma.Wm82.gnm4.ann1.Glyma.01G035000, glyma.Wm82.gnm4.ann1.Glyma.01G041400, glyma.Wm82.gnm4.ann1.Glyma.01G041450, glyma.Wm82.gnm4.ann1.Glyma.01G042100, glyma.Wm82.gnm4.ann1.Glyma.01G081600, glyma.Wm82.gnm4.ann1.Glyma.01G081700, glyma.Wm82.gnm4.ann1.Glyma.01G105000, glyma.Wm82.gnm4.ann1.Glyma.01G112500, glyma.Wm82.gnm4.ann1.Glyma.01G113400
Results for List 1, showing `biological_process`
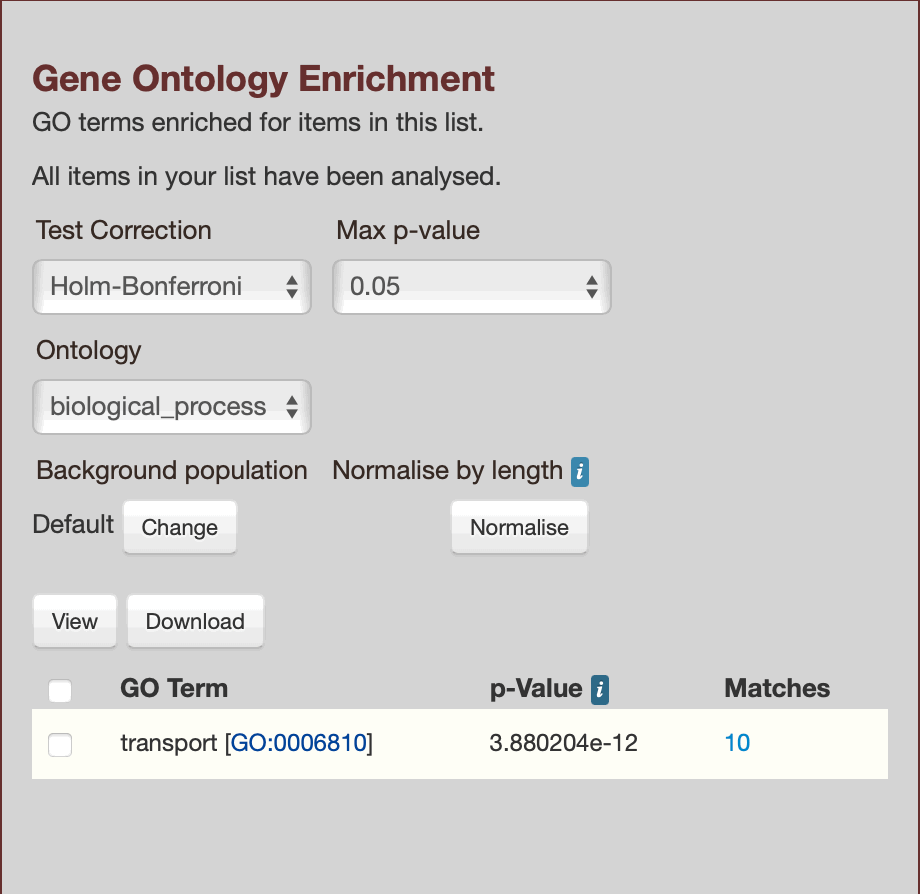
glyma.Wm82.gnm4.ann1.Glyma.01G128700, glyma.Wm82.gnm4.ann1.Glyma.01G155300, glyma.Wm82.gnm4.ann1.Glyma.03G041600, glyma.Wm82.gnm4.ann1.Glyma.03G146200, glyma.Wm82.gnm4.ann1.Glyma.05G158300, glyma.Wm82.gnm4.ann1.Glyma.08G116000, glyma.Wm82.gnm4.ann1.Glyma.09G194900, glyma.Wm82.gnm4.ann1.Glyma.09G279900, glyma.Wm82.gnm4.ann1.Glyma.11G089400, glyma.Wm82.gnm4.ann1.Glyma.13G162500
Results for List 2, showing `cellular_component`
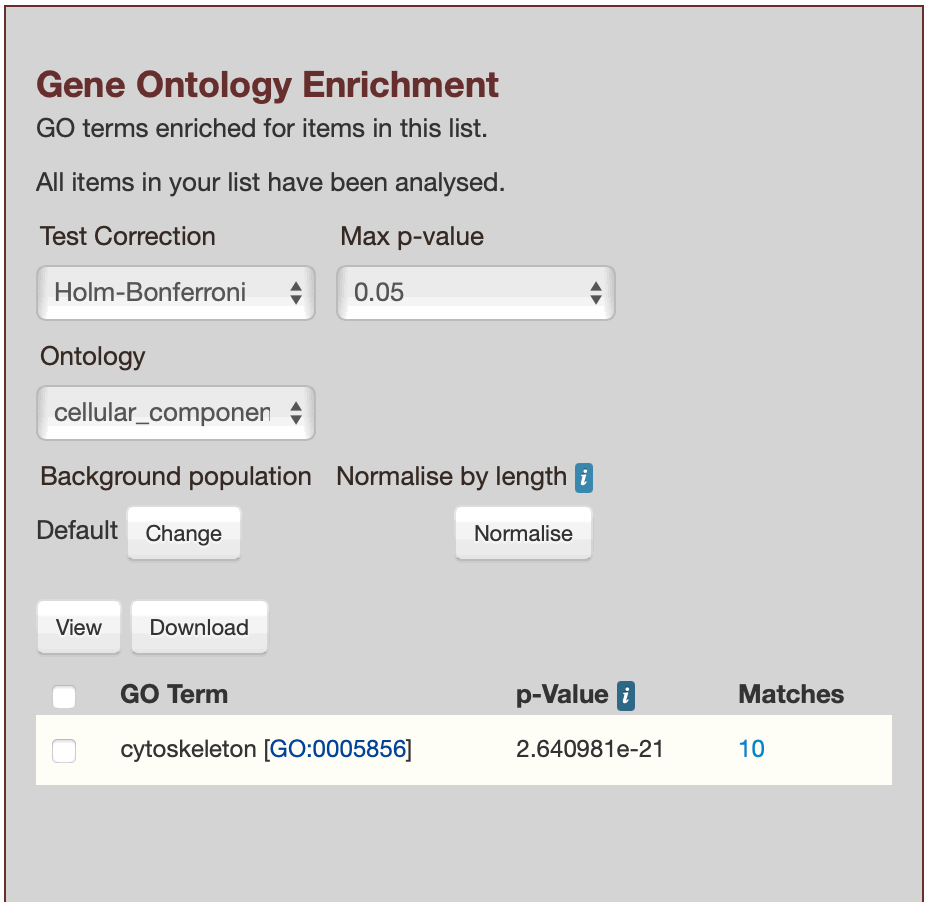
glyma.Wm82.gnm4.ann1.Glyma.01G032400, glyma.Wm82.gnm4.ann1.Glyma.01G032900, glyma.Wm82.gnm4.ann1.Glyma.01G033200, glyma.Wm82.gnm4.ann1.Glyma.01G033300, glyma.Wm82.gnm4.ann1.Glyma.01G039000, glyma.Wm82.gnm4.ann1.Glyma.01G046900, glyma.Wm82.gnm4.ann1.Glyma.01G060300, glyma.Wm82.gnm4.ann1.Glyma.01G112200, glyma.Wm82.gnm4.ann1.Glyma.01G112300, glyma.Wm82.gnm4.ann1.Glyma.01G125300
Results for List 3, showing `molecular_function`